Home » Products » Chromatography/Mass Spectrometry » MALDI-Based Instruments and Solutions » Shimadzu Microbial Identification Solutions
Microbial Identification Solutions
Specialized High-Throughput Microbial Identification
Shimadzu Microbial Identification Solutions
Shimadzu’s analytical philosophy is technically centered on the integration of "Analytical Intelligence" across its entire portfolio of chromatography and mass spectrometry systems. This approach represents a shift from manual instrument management to an automated, self-optimizing laboratory ecosystem.
Technically, this is achieved through hardware that features automated system checks, remote monitoring via mobile platforms, and self-healing vacuum systems that reduce maintenance intervals. On the software side, the inclusion of deep-learning AI engines like "Peakintelligence" allows for the processing of vast datasets with the objective precision of an expert scientist.
Rapid Three-Step Identification Workflow
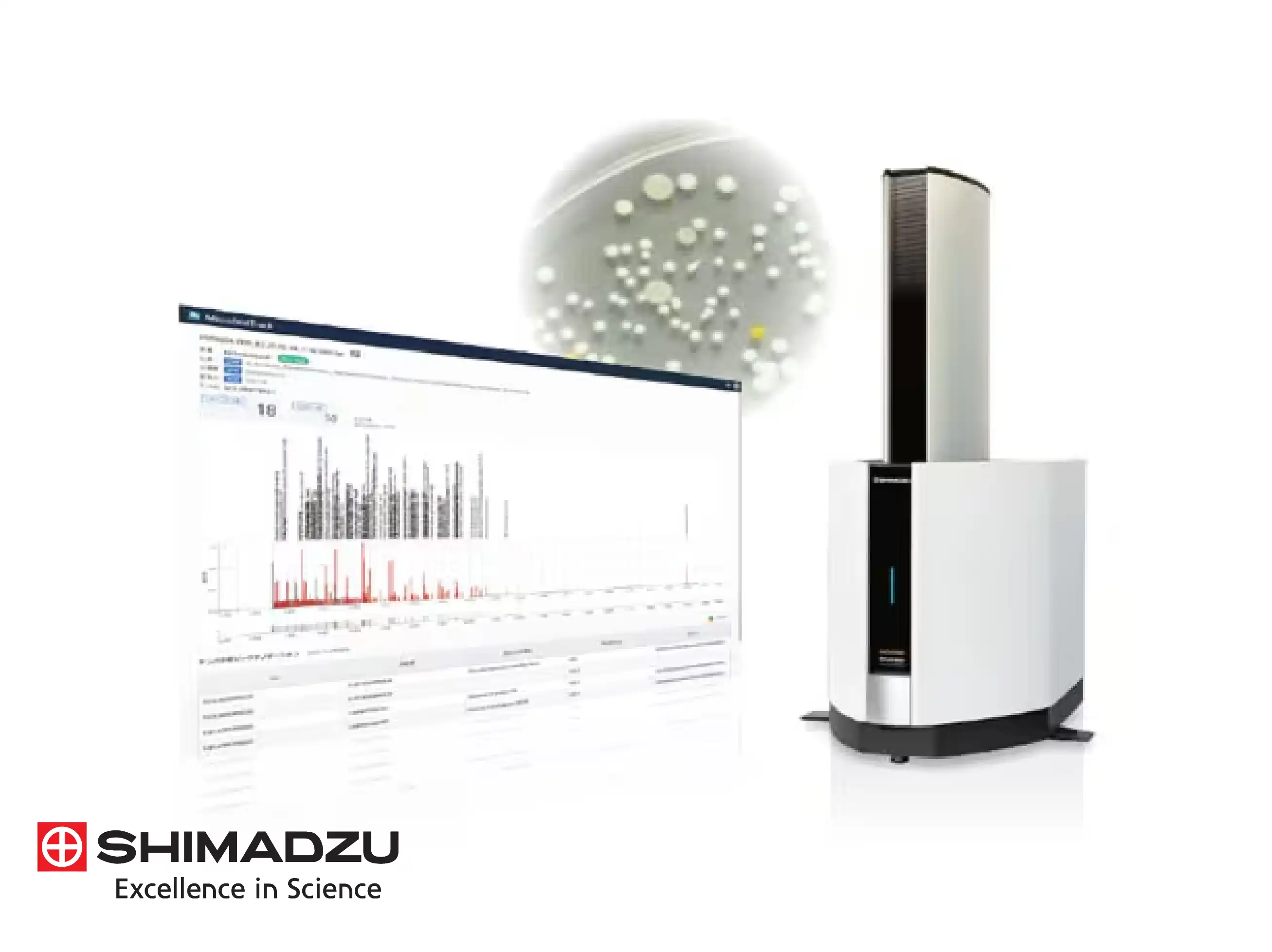
The Shimadzu microbial identification system is technically designed for maximum efficiency through a streamlined three-step process: sample preparation, measurement, and database matching. Analysts simply apply a small amount of a colony directly onto a disposable MALDI plate, add a matrix reagent, and introduce it to the mass spectrometer. The system then automatically acquires a unique mass spectrum for the species and compares it against validated databases, delivering final results in under two minutes per sample.
Genomic-Based Theoretical Protein Database (MicrobialTrack)
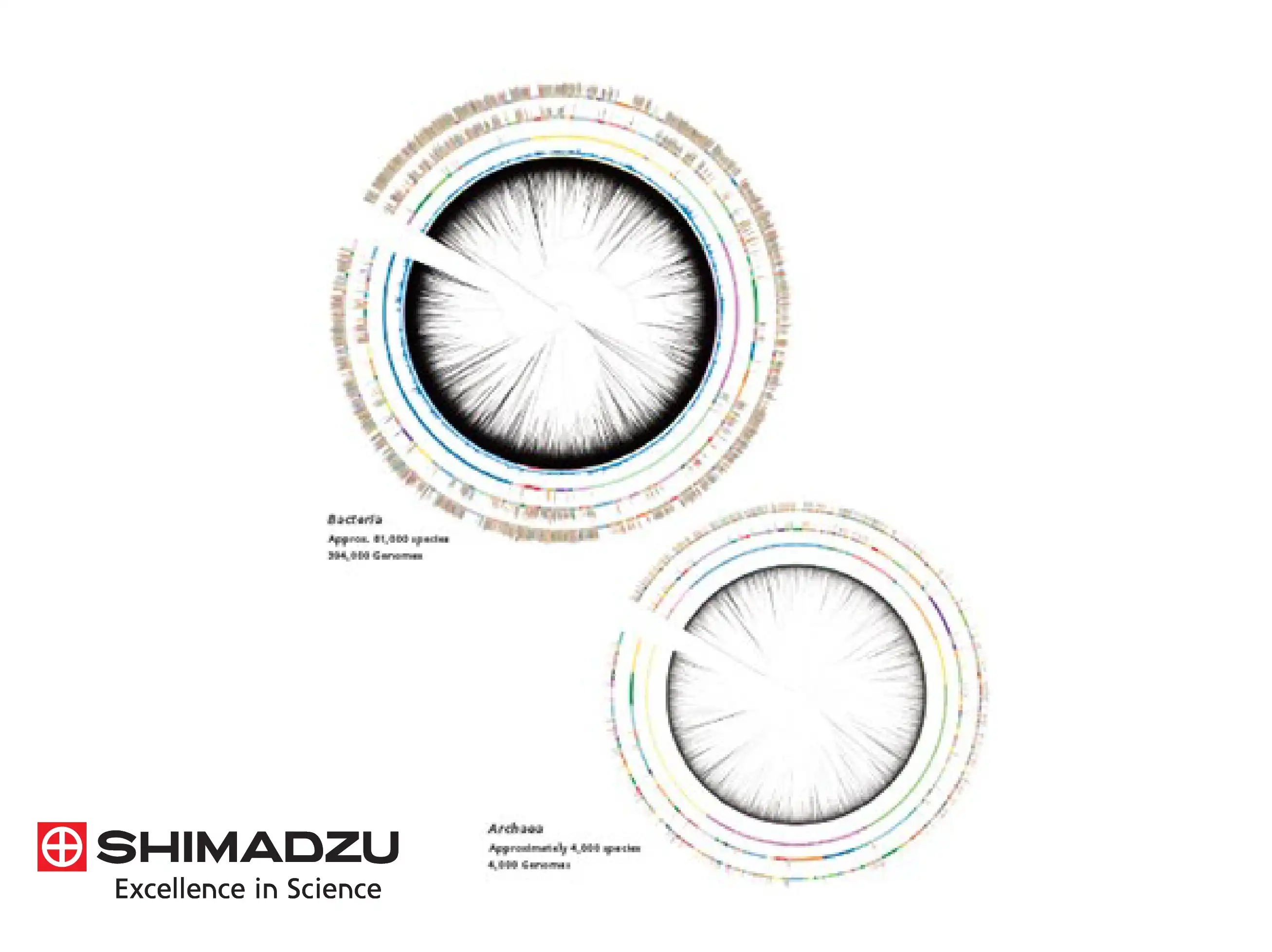
A significant technical innovation is the use of MicrobialTrack, an analytical platform that utilizes a vast database of theoretical protein masses predicted directly from genomic sequences. Unlike conventional fingerprinting which relies purely on observed spectra, this approach enables the identification of approximately 18,000 prokaryotic species with valid names and over 85,000 species predicted from genomic data. This allows for the accurate classification of microorganisms that have never been cultured in a laboratory environment.
High-Precision Ribosomal Protein Fingerprinting
Technically, the identification is based on the detection of ribosomal proteins, which account for nearly 20% of total protein expression in the logarithmic growth phase. Because these proteins are highly conserved and easily ionized due to their basic nature, they serve as stable markers that are minimally influenced by environmental conditions or culture media. The software extracts these marker peaks to create a robust spectral signature that remains consistent across different laboratory settings.
Advanced Strain-Level Discrimination
For applications requiring more than just species-level identification, Shimadzu offers "Strain Solution" software. This technical extension utilizes the S10-GERMS method, which links ribosomal protein molecular weight information to specific genetic sequences. By pre-registering unique markers, the system can differentiate between genetically similar bacteria, subspecies, and specific strains, making it a critical tool for epidemiological tracking, contamination source investigation, and library maintenance.
SuperSpectra for Enhanced Robustness
The systems utilize SuperSpectra technology to reduce the incidence of false positives. SuperSpectra are technically curated groups of peak markers extracted from at least 15 mass spectra per strain, obtained under varying measurement conditions across multiple laboratories. This ensures that the identification algorithm focuses on stable, reproducible signals, overcoming the historical challenge of mass-spectrum variability and ensuring reliable results even for complex or poorly prepared samples.
High-Throughput Clinical and Industrial Capacity
Designed for high-demand environments, the system can process up to 380 samples within five hours in high-throughput mode. This efficiency is supported by automated ion source cleaning and autotuning features (EasyCare), which maintain optimal instrument performance without requiring specialized technical expertise. This high capacity makes it ideal for clinical diagnostics, food safety monitoring, and drug discovery processes where rapid turnaround is essential for managing toxic microorganisms or infectious pathogens.
Open and Customizable Database Architecture
The software provides an open system architecture, allowing users to register new entries or create entirely custom databases. Technically, this enables researchers to build proprietary libraries for niche environmental samples, specific industrial contaminants, or newly discovered effective microorganisms in drug discovery. The ability to perform molecular profiling and clustering allows for tracking the evolution of microbial communities over time in a secure, networked data environment.
Vendor-Neutral and Cloud-Accessible Analysis
MicrobialTrack is technically vendor-neutral, supporting universal data formats from various MALDI-TOF models. Furthermore, the platform is accessible via cloud services, ensuring that users always have access to the latest database versions without manual updates. This technical synergy allows for multifaceted evaluation metrics, such as random resampling to estimate identification certainty levels, providing analysts with a clear confidence score for every genus and species identified.
Click here for more information about Shimadzu range of products
Related Products
CONTACT US
CONTACT US
Taawon for Laboratory & Scientific Supplies
Taawon Group © 2026 - All Rights Reserved
We use cookies to optimize site functionality and give you the best possible experience